A 10 year anniversary: pioneering GPU computing
Anniversaries are nice, especially in December! So here is one to share with you all: it has been 10 years since Anatoli, Heather, and Max (the 3 co-founders of Neurala) begun tinkering with GPUs and Deep Learning, setting the foundations of Neurala while getting their Ph.D at Boston University. This post is a re-publication of an earlier post based on our work on AMD GPUs. Funny to say, much of the techniques, which are now in an awarded patent, are still the backbones of Neurala's tech. But let's get back to the past!
By Anatoli Gorchetchnikov, Heather Ames, Massimiliano Versace
The last post on GPU made me think of a project Anatoli Gorchetchnikov, Heather Ames and myself embarked on in 2006 when we got really interested in general purpose computing on graphic processing cards. At the time, there was no CUDA or OpenGL available: programming GPUs was really tough. But we tried, with some very good results, to port some of the models we used on GPUs. Here is how we did it.
First of all, a disclaimer: the work refers to 2006, and much has changed since then in the way you program GPUs. Also, the GeForceR 6800 used at the time is a museum piece…nevertheless, many of the arguments that follow are still relevant.
As you know, artificial neural networks (ANNs) are highly parallel, computationally intensive mathematical models that capture the dynamics and adaptability of biological neurons. Many of these networks, especially those that are biologically inspired, are dynamical systems requiring numerical integration to calculate their variables at every moment of time as a function of previous values of one or more of these variables. In certain classes of ANNs, neurons are described in terms of complex system of differential equations, and a significant degree of recurrency can characterize their connectivity matrix. Graphic processing units (GPUs), originally designed for computer gaming and 3D graphic processing, have been used in general-purpose, computationally intensive models in application domains ranging from physical modeling, image processing, and computational intelligence. Biologically realistic neural networks are used to model brain areas responsible for perceptual, cognitive, and memory functions. For instance, one of us, Anatoli Gorchetchnikov, has done considerable work in using biophysically realistic neural models of cell dynamics and learning laws to explain how rats learn to navigate novel environments (Gorchetchnikov and Hasselmo, 2005a,b; Gorchetchnikov at al., 2005a,b). As time progresses, these models tend to become increasingly realistic, and require more computing power to run. Advances in graphics hardware and software have enabled a wider array of algorithms to be ported on GPUs, with considerable increase in execution speed with respect to traditional CPU programming.
This post discusses an implementation of Recurrent Competitive Field (RCF) (Ellias and Grossberg, 1975) and Hodgkin-Huxley (Hodgkin and Huxley, 1952) neural networks on a NVidia GeForce 6800 Ultra GPU. These models are used in a variety of theoretical and applied study. Porting them to GPU presents some challenges. Two critical issues that have to be addressed when implementing large-scale ANNs on GPU: the presence of extensive or all-to-all connectivity between processing elements, and computational precision when integration of complex differential equations is required. Performance analysis in comparison with CPU implementations shows considerable gain for the GPU versions of these networks even when the output precision is matched with the precision of the integration shown by the CPU versions. This analysis also pinpoints several limitations of GPU implementations of large ANNs. One key aspect of these implementations has not been fully addressed previously due to the difficulties in achieving recurrent interdependencies in streamed architecture.
Why do we need GPUs?
Computational neuroscience uses neural models to investigate the properties and functionality of the nervous system. These models range in complexity and level of biological detail from simple autoassociative memories implemented with a single differential equation per neuron to very detailed multicompartmental models that use thousands of equations per neuron. Nevertheless, all these models share a common trait. They are usually either hard to analyze or even mathematically intractable and require numerical simulations to achieve results showing changes of neural activity over time. At the same time, neural models are inherently parallel and allow a simple mapping on single instruction multiple data (SIMD) computational paradigm.
A variety of specialized software packages is available today to support large-scale ANNs simulations. Among others, NEURON (Hines 1989, 1993; Hines and Carnevale 1994; Carnevale and Hines 2006), GENESIS (Bower and Beeman 1998), KInNeSS (Versace et al., 2008), CSIM (Maass et al. 2002; Natschläger et al. 2002), NEST (Gewaltig and Diesmann 2007) and SPLIT (Hammarlund and Ekeberg 1998). An excellent review of currently available neural simulation packages can be found in Brette et al. (2007). Several of these software packages can be run on parallel hardware to allow for larger simulations. NEURON can be run on parallel hardware including Beowulf clusters, the IBM Blue Gene and Cray XT3. Parallel GENESIS (PGENESIS) runs on almost any parallel cluster, SMP, supercomputer, or network of workstations where MPI and/or PVM is supported. NEST supports distribution of a simulation over multiple processors of an SMP machine or over multiple machines in a cluster. Both Split and NCS can be run on clusters-of-workstations. However, the availability and cost of clusters of PC and supercomputers limits the access to massively parallel hardware for the modeler.
The evolution of programmable graphic processing units (GPUs) offers increasing opportunity to use GPU parallel SIMD architecture for non-graphic computations, such as large-scale ANNs simulations. GPUs are modern, powerful programmable processors that are increasingly used for general-purpose computing. The term General-Purpose GPU (GPGPU) computing refers to the large community of researchers and programmers that develop GPU applications outside the traditional domain of computer graphics. GPGPU’s homepage http://www.gpgpu.org/ provides a list of recent applications and valuable resources. High level languages have emerged in the past few years that have provided GPGPU programmers with more sophisticated tools that support more complex operations. HLSL, Cg, Ashli, Sh, OpenGL Shading Language, CUDA, and OpenCL are examples of languages that allow developers to write GPU programs in a C-C++ programming style. The introduction of double precision will favor a more widespread use of GPUs in very large scale scientific applications in the computational sciences. CPUs and GPU have a fundamentally different architecture which accounts for the different applications domains that these processors are best suited for. CPUs are more efficient in handling sequential-style processing due to the fact that many of the transistors on the CPU are dedicated to tasks such as caching and branch prediction. GPUs, on the other hand, are more suitable for highly intensive arithmetic operations due to the large availability of transistors that can be dedicated to these demanding computations.
GPUs is a highly specialized chips built for accelerating computer graphics applications, and many obstacles still remains to allow programmers to fully exploit the high resources of GPUs for GPGPU applications. Despite these obstacles, the faster pace of development of GPUs with respect to CPUs, the increasing availability of a broad array of languages that assist the GPU programmer, and the fact that the graphic processor is a very powerful and affordable computational hardware are important benefits that justify the effort of programming a graphic processor for non-graphical applications
Biophysically realistic artificial neurons are particularly suited for GPU implementation. Neurons whose membrane potential are described in terms of differential equations represent a special case of a larger set of operations that can be effectively parallelized on GPUs. Fortunately, GPUs have evolved over the years to handle complex physics simulations in the context of gaming. Ordinary differential equations and partial differential equations have been solved on GPUs in applications ranging from particle systems simulations (Kipfer et al., 2004), gravitational force computations (Nylander et al, ?), particle collision simulations (Kolb et al, 2004), and heat equation (Rumpf and Strzodka, 2005). Hagen et al [2005] modeled the shallow water wave dynamics using a system of SaintVenant equations. They used second order Runge-Kutta method for numerical integration and achieved about 25-fold increase of speed of the GPU version of the solution over the CPU version. Georgii and Westermann [2005] simulated mass-spring system integrated through time.
Artificial neural networks have also been simulated on GPU. Bohn et al. (1998) have implemented Kohonen maps in graphic hardware. Several attempts to implement traditional artificial neural networks (ANNs) using GPU were undertaken recently. Rolfes (2004) implemented GPU version of the multi-layer perceptron through DirectXR. Oh and Jung (2004) used the same approach to speed up the matrix multiplication in back propagation neural network (another name for a multi-layer perceptron). Similar approach was used by Ilie (2002). Davis (2005) pointed out that approach of these authors requires batch processing and is not applicable to ANNs architectures where the input changes on trial-by-trial basis. Davis (2005) conducted the investigation of back propagation, Hopfield network (Hopfield, 1982), and fuzzy Adaptive Resonance Theory (ART, Carpenter et al., 1991) on GPU. While his results for the first two networks showed considerable performance gain, his results for fuzzy ART on GPU were worse than CPU. This was due to constant bus traffic that was necessary to support an on-line learning algorithm of ART. Davis pointed out that this difficulty can be overcome by using the OpenGLR render-to-texture extensions, which were not implemented at the time of his investigation [Davis 2005]. Meuth and Wunsch (2007) have implemented ART networks on GPU achieving encouraging results. Bernard and Keriven (2005) provided first insights on using GPU for biologically realistic spiking neural networks. Their solution is very similar to the one proposed here, but the authors did not concentrate on the issues that are investigated here.
ANNs on GPUs?
The level of complexity of neural models comes from several factors. One is the complexity of the connectivity between neurons. Extensive connectivity with dynamic weights that are updated through learning algorithms requires keeping all synaptic weights in memory. With a sufficiently large network and extensive connectivity, the memory capacity of the system can be easily exhausted. One of the issues that was investigated in this earlier work was whether the extent of the connectivity matrix will decrease the applicability of GPGPU to neural models. Dynamic recurrent competitive field network (RCF, Ellias and Grossberg 1975) with full connectivity was implemented to answer this question. The second issue addressed here is the precision of computation. Hodgkin-Huxley equations (Hodgkin and Huxley, 1952) contain fast variables and require careful selection of the integration step in order to achieve sufficient precision. CPU implementations of these equations usually build upon double precision floating point numbers to increase the value of integration step without the loss of precision of the results. GPU implementation of integrate-and-fire neurons do not suffer from this precision issue (Bernard and Keriven, 2005) since these equations do not have fast variables, and do not suffer from precision issues due to it slow dynamics. However, more accurate neural models require full Hodgkin-Huxley dynamics. Current GPU architectures only allow single precision floating point computations, and therefore would require shorter integration step to achieve the same resulting precision. Shorter step means more steps for the same simulated time interval, and this can nullify the effect achieved by parallel processing on the GPU. A network of Hodgkin-Huxley neurons was implemented here to investigate this issue.
Background and Implementation
We used the developmental version of Synchronous Artificial Neuronal Network Distributed Runtime Algorithm (SANNDRA) library. The previous version of SANNDRA is a part of the KDE Integrated NeuroSimulation System (KInNeSS) - an open source software for general purpose neural simulations available at www.kinness.net. KInNeSS a neurosimulation software best suited to simulate networks of hundreds to thousands of branched multi-compartmental neurons with biophysical properties such as membrane potential, voltage-gated and ligand-gated channels, the presence of gap junctions or ionic diffusion, neuromodulation channel gating, the mechanism for habituative or depressive synapses, axonal delays, and synaptic plasticity (Versace et al., 2008). Since KInNeSS is mainly a graphic user interface and visualization tool built on top of the computational engine of SANNDRA and the main issues studied in this paper are computational performance and precision, we bypassed KInNeSS in our simulation and used a command line application based directly on SANNDRA. SANNDRA was originally designed in 1997–9 for MasPar MP1 parallel computer with SIMD architecture and ported for Linux as a partial SIMD emulation in 2000. It was intended to run a long iterative loop through relatively simple computation done in parallel on many similar elements. Later design in 2001–5 relaxed SIMD requirements on the similarity of computational elements, but these elements are still synchronized for the data exchange. On a sequential computer this leads to an unavoidable performance loss due to synchronization, but ensures that these elements get correct input signals. Each element can have an access to the output of any other element. This design makes SANNDRA capable of numerical integration of large systems of non-homogeneous differential equations, although it can be put to much simpler use like iterative solving systems of algebraic equations or image processing, when each element represents a pixel. SANNDRA version 1.0.0 available as a snapshot and a current version 1.2.0 available through anonymous CVS were designed as a single module. Currently under development is version 2.0.0 that will be licensed under LGPL. This version is used to perform the following simulations. Two ANNs models are implemented in this work: Recurrent Competitive Fields (Ellias and Grossberg, 1975) and a network of Hodgkin-Huxley neurons (Hodgkin and Huxley, 1952). The following section describes the numerical integration technique used to implement these networks.
Numerical Integration on GPU
The activation of each neuron was stored in the NVidia GeForce 6800 Ultra GPU texture with 32-bit floating point precision. Since there are four colors per pixel, each pixel in the texture contained four neuronal activations. As a result, the dimensions of the texture were half of the corresponding network dimension (rounded up in case of odd network dimension). Since GeForce R 6800 Ultra has 16 fragment pipelines, this packing scheme allows simultaneous processing of 64 neurons on every stage of these pipelines. Standard fourth order Runge-Kutta method was used to calculate the derivative of neuronal activation on each integration step. All four substeps of this method were implemented in a fragment shader within a single rendering pass. One of the concerns with numeric simulations on GPU is that results often need to be transferred back to the CPU and saved on disk for further analysis off line. On the other hand, using render to texture functionality does not require this back transfer for the simulation itself to proceed correctly. Therefore, two versions of implementation were provided. One transferred the results back to CPU on every time step using glGetTexImage call, the other skipped this transfer. This allowed for the analysis of the possible slowdown caused by the back transfer.
Recurrent Competitive Field
Ellias and Grossberg (1975) discussed a neural architecture called recurrent competitive field (RCF). RCFs are a mathematical abstraction of ubiquitous on-center/off-surround neural architecture found in the mammalian brain, and have been widely used in modeling cognitive, behavioral, and physiological data This network is described by a system of differential equations
![]()
where: xi(t) is activity in the i-th node, A, B, C and D – parameters. The behavior of this network depends on two factors: whether BC/A is greater than 1 and whether C/D is greater, equal or less than 1. The former ratio defines whether the activation of the network will persist over time or decay to 0. The latter ratio defines whether the initial activation pattern will be uniformized, preserved, or “contrast enhanced” up to a winner-take-all final state. From the perspective of this study, RCF is an interesting test case because each neuron has only one variable, but requires inputs from all other neurons. To query all these inputs a neuron has to loop through them, and GeForceR 6800 Ultra that was used here allows the maximal loop size of 256. In two dimensions it limits the access to inputs with the square of 256x256 pixels. Even when neurons are packed four into pixel as was done here, this limits the network size to 512x512 neurons. Bernard and Keriven (2005) avoided this issue since they used spiking neurons and only needed to count the number of neurons that produced a spike at every time step. In the networks with analogous inputs this solution is not applicable. Fragment feedback loop based on render-to-texture OpenGLR extension simplifies the implementation of neural interactions through writing output values into textures that serve as an input on the next simulation step. Subsequent releases of NVidiaR drivers with render-to-texture capability allowed extensive testing of neural models on programmable graphics hardware. Equation (1) was implemented in a fragment shader using GLSL. Parameters A, B, C, and D were hardcoded in the shader as constants, but can be easily passed as uniform variables. Note, that Equation (1) can be rewritten so that the sum of all inputs needs to be calculated instead of the sum in the equation. In this case there would be no need to calculate this sum in every fragment, it can be calculated either once on the CPU or once per vertex in the vertex shader and passed to a fragment shader. This will significantly improve the performance. However, the purpose of this study was not to optimize the specific performance of RCF network, but to investigate the limitations of extensive texture lookups. In the case of any kind of learning in these projections, the weight matrix for each neuron will quickly become unique and this situation would not allow the sum optimization. To appreciate this future problem, optimization of the sum was avoided in both GPU and CPU implementations. Without it the number of texture lookups per neuron per iteration is equal to the number of neurons N, and the complexity of the whole problem is O(N2).
Hodgkin-Huxley Equations
Hodgkin-Huxley equations (Hodgkin and Huxley 1952) were designed to approximate the spread of an action potential (spike) through the giant squid axon. They are based on the Kirchhoff’s current law and contain voltage dependent sodium and potassium currents as well as leakage current and optional current injection
where A, B, and V0 are parameters. Hodgkin and Huxley (1952) used am and an as linoid functions; bm, bn, and ah as exponential functions; and bh as sigmoid function for their squid axon simulations.
Subsequent research has shown that similar formalism with some parameter changes and rate function rearrangement can describe many more other types of neurons. For the simulations presented here the original set of parameters (Hodgkin and Huxley 1952) was used. The whole set of equations presented in this section was implemented using four fragment shaders. Each of the variables Vm, m, h, and n had a designated primitive and a designated shader. The order of rendering was not important, since on every rendering pass the results for each variable were calculated using a set of input textures that resulted from the previous time step and written into a set of output textures. After the step was completed the input and output textures were swapped by reassigning the corresponding uniforms and binding the former input textures as output. This method of swapping avoided the expensive texture copying. For each neuron the number of texture accesses was fixed: five to calculate the voltage (previous voltage, m, h, n, and current injection J) and two for each of the gating variables (previous values of the variable and voltage). This added up to total of eleven texture accesses per neuron per iteration. For simplicity, there was no connections between neurons in this “network”, and the complexity of the task scaled linearly (O(N)) with the number of neurons N.
All simulations were performed using dual OpteronR 240 workstation with 2Gb of RAM and AGP-8X NVidiaR GeForce 6800 Ultra with 256Mb of video RAM. Operating system was based on SuSER Linux 9.2 with the custom kernel 2.6.14.3 and NVidiaR OpenGLR drivers version 81.74 for 64-bit systems. The integration step was set to 0.05 (arbitrary time units for RCF, simulated mS for Hodgkin-Huxley) in all CPU and GPU simulations except the cases explicitly discussed below. CPU simulations were set to run with either one computational thread or with two threads to utilize the dual processor architecture. In both CPU and GPU simulations the disk output was performed in a separate thread. Simulation time was measured for the computational thread(s) wall clock execution. Since OpenGLR context has to be initialized and used within the same thread, for GPU runs the simulation time includes the cost of context initialization and cleanup as well as shader compilation time. Since most of neural simulations work with populations of neurons that are not squares, the networks in this study also used rectangular populations. In the results below only the total number of neurons in the network is reported.
Results
Recurrent Competitive Field
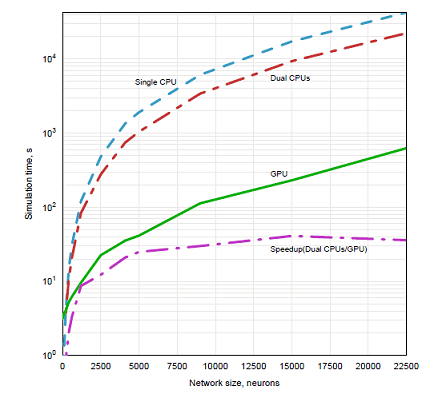
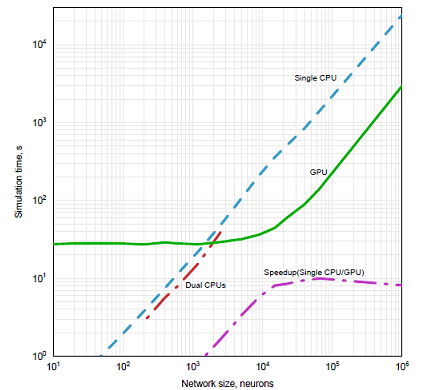
Figure 1: Performance graph for RCF.
Results for the RCF simulations are presented in Figure 1. The total simulation length for these runs was 60000 integration steps. The maximal speedup of GPU implementation over dual CPU implementation was 41-fold for the network size of 15000 neurons. The major concern about this set of simulations was whether the limited number of iterations in the loops within a fragment shader will limit the size of networks with all-to-all connectivity that can be implemented on the GPU. This was not the case, since another bottleneck happens before the maximal loop size is reached. One of the CPUs in the system was loaded by running the simulation. For the three largest networks simulated (sizes 9000, 15000, and 22500 neurons), an increasing load of X-Windows system was observed on the second CPU (up to 60% in the case of 22500 neurons). This load pushed the system into a sluggish irresponsive to user input state and made the system unusable during the simulation. Another result of this load is the decrease in speedup between 15000 and 22500 neurons observed in Figure 1. The second concern was the cost of transferring data back to the CPU, especially considering that the test system used AGP and not PCI-Express. This was not an issue with these simulations. The total simulation time with and without back transfer did not differ by more than 5% (e.g for the network size of 4000 neurons the time increased from 34.1s to 35.5s). The bottleneck happened when these data were actually written to disk (for the same network the simulation time increased from 35.5s to 77.4s). Note, that this bottleneck is not related to GPU, the same network size on the CPU also takes extra 20–30s to output the results to disk, except that for the total running time of 747s this increase does not appear as crucial as for the GPU version. The absolute time of disk output for CPU version is no shorter than for GPU implementation, but since disk operations are performed in a separate thread during the computation, the large the computational load on CPU, the more of disk I/O time is hidden. The precision of GPU implementation was not a concern, since RCF has a slow dynamics. The results confirmed this, since the difference between double precision CPU version and single precision GPU version only showed in the sixth significant digit. These results support the claim that there is no precision issues with the leaky integrate-and-fire (IAF) neurons (Bernhard and Keriven 2005), since leaky IAF also has slow dynamics similar to Equation (1).
Hodgkin-Huxley Equations
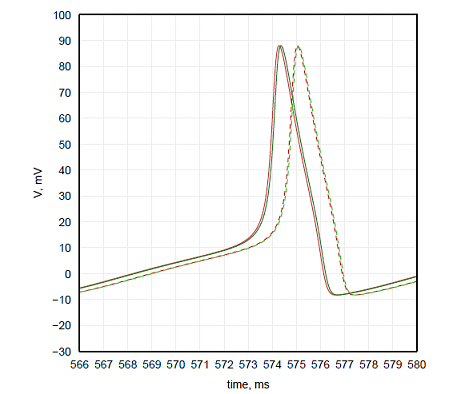
With Hodgkin-Huxley equations the precision of the integration was the primary concern. Since these equations have fast dynamics there is a larger opportunity for error to occur, and since it is an initial value problem, this error will accumulate over time. To consider how large the error is the simulation was ran for 600 simulated milliseconds. The integration step was gradually increased for the CPU implementation from 0.001ms until the difference in spike timing for the last spikes in the simulation was greater than 0.1ms for the spikes at the end of simulation. This happened for the time step of 0.02ms, so the time step of 0.01ms was used as a baseline for CPU implementation. Further increase of the time step shown that the values 0.05ms and smaller lead to the difference of spike timing from the baseline that is below 1ms, which is usually a sufficient precision for neural simulations. Baseline with time step 0.01ms and the results with time step of 0.05ms are plotted in red in Figure 3.2.
Figure 2: Voltage traces of Hodgkin-Huxley equations with different integration steps. Solid traces represent baselines, dashed lines represent acceptable error. CPU results in red, GPU results in green.
GPU implementation uses single precision floating point numbers instead of double precision used by
CPU. As a result even the baseline that was achieved at the time step of 0.001ms was slightly shifted from CPU baseline. Using the same criteria as above, the acceptable time step for GPU implementation was found to be 0.004ms. The results of these simulations are plotted in Figure 2 in green.
Figure 3: Performance results for Hodgkin-Huxley equations.
The conclusion was that to achieve the same precision the integration time step for GPU implementation
should be 12.5 times shorter than for CPU version. The rest of the simulations ran for 250 simulated ms and with time steps 0.05ms for the CPU version and 0.004ms for the GPU version. Performance results for these runs are shown in Figure 2. These results show an order of magnitude speedup of GPU implementation over CPU version for sufficiently large networks despite matching precision of the integration. The flat segment of the results for GPU simulations of small networks is partially due to the constant time necessary to initialize the context and to compile four shaders. Another reason for this segment to be flat is that GPU resources are not fully utilized for small networks.
Conclusions
The results presented here, despite referring to an older system, can be of general interest if one wants to implement large scale neural simulations on GPU. Some of the issues we refer below have been solved with more modern processors/languages, but I will present them as they were formulated… for archeological interest!
The first is, of course, the issue of exhausting the video memory with matrices of connection weights. The second problem is overloading of underlying windowing system with excessive texture lookups necessary to access the outputs of other neurons in the network. This issue can have several solutions. One solution is to dedicate the multiprocessor computational server for the simulations and not to use windowing system on this server for anything other than subserving GPU simulations. Another and much better solution will be available as soon as optimized image processing calls will be available in shading language. A lot of neural connectivity kernels are Gaussian or similar type kernels that are also used in image processing. Since neuronal activity is represented as a texture, getting input through such a kernel will be virtually identical to applying Gaussian filter to an image. For all-to-all connectivity, mipmapping of the input texture down to size 1x1 also could solve the problem if it would be possible on 32-bit floating point precision textures. The third problem is the reduced precision of the GPU (single precision floating point) relative to double precision floating point commonly used for CPU simulations. As the results here show, this only becomes an issue for differential equations with fast dynamics, and it can be corrected by reducing the integration step while still keeping some performance gain. Introduction of 64-bit precision on the GPU will immediately allow speeding up simulations of systems with fast dynamics by an order of magnitude just due to the increase in integration step.
Aside from these issues, there is an obvious advantage of using GPU to speed up large scale neuronal simulations. Rapid innovation in the GPU hardware and software industry, as well as in the GPGPU community, this will open a set of new possibilities for computational neuroscience who seek alternative solutions to PC clusters and supercomputers to run massively parallel neural simulations on commoditized hardware.
References
BERNHARD, F., AND KERIVEN, R. 2005. Spiking neurons on GPUs. Tech. Rep. 05-15, Ecole Nationale des Ponts et Chauss´es.
CARPENTER, G. A., GROSSBERG, S., AND ROSEN, D. B. 1991. Fuzzy ART: Fast stable learning and categorization of analog patterns by an adaptive resonance system. Neural Netw 4, 759–771.
DAVIS, C. E. 2005. Graphic Processing Unit Computation of Neural Networks. Master’s thesis, University of New Mexico, Albuquerque, NM.
ELLIAS, S. A., AND GROSSBERG, S. 1975. Pattern formation, contrast control and oscillations in the short term memory of shunting on-center off-surround networks. Biol Cybern 20, 69–98.
GEORGII, J., AND WESTERMANN, R. 2005. Mass-spring systems on the GPU. Simulation Modelling Practice and Theory 13, 693–702.
GORCHETCHNIKOV, A. 2000. An Approach to a Biologically Realistic Simulation of Natural Memory. Master’s thesis, Middle Tennessee State University, Murfreesboro, TN.
GORCHETCHNIKOV A., VERSACE M., HASSELMO M. E. (2005a). A Model of STDP Based on Spatially and Temporally Local Information: Derivation and Combination with Gated Decay. Neural Networks, 18, 458-466.
GORCHETCHNIKOV A., VERSACE M., HASSELMO M. E. (2005b). Spatially and temporally local spike-timing-dependent plasticity rule. In: Proceedings of the International Joint Conference on Neural Networks, number 1568 in IEEE CD-ROM Catalog Number: 05CH37662C, pp. 390-396.
GORCHETCHNIKOV A., HASSELMO M. E. (2005a). A simple rule for spike-timing-dependent plasticity: local influence of AHP current. Neurocomputing, 65-66, 885-890.
GORCHETCHNIKOV A., HASSELMO M. E. (2005b). A biophysical implementation of a bidirectional graph search algorithm to solve multiple goal navigation tasks. Connection Science, 17(1-2), 145-166.
HAGEN, T. R., HJELMERVIK, J., LIE, K.-A., NATVIG, J., AND OFSTAD HENRIKSEN, M. 2005. Visual simulation of shallow-water waves. Simulation Modelling Practice and Theory 13, 716–726.
HODGKIN, A. L., AND HUXLEY, A. F. 1952. Quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol 117, 500–544.
HOPFIELD, J. 1982. Neural networks and physical systems with emergent collective computational abilities. In Proc Natl Acad Sci USA, vol. 79, 2554–2558.
ILIE, A. 2002. Optical character recognition on graphics hardware. Tech. Rep. integrative paper, UNCCH, Department of Computer Science.
OH, K.-S., AND JUNG, K. 2004. GPU implementation of neural networks. Pattern Recognition 37, 1311–1314.
ROLFES, T. 2004. Neural networks on programmable graphics hardware. In Game Programming Gems 4, A. Kirmse, Ed. Charles River Media, Hingham, MA, 373–378. 13
MEUTH, J.R. and WUNSCH, D.C. (2007) A SURVEY OF NEURAL COMPUTATION ON GRAPHICS PROCESSING HARDWARE. 22nd IEEE International Symposium on Intelligent Control, Part of IEEE Multi-conference on Systems and Control, Singapore, 1-3 October 2007
KIPFER, P., SEGAL, M., and WESTERMANN, R. 2004. "UberFlow: A GPU-Based Particle Engine." In Proceedings of the SIGGRAPH/Eurographics Workshop on Graphics Hardware 2004, pp. 115–122.
KOLB, A., L. LATTA, and C. REZK-SALAMA. 2004. "Hardware-Based Simulation and Collision Detection for Large Particle Systems." In Proceedings of the SIGGRAPH/Eurographics Workshop on Graphics Hardware 2004, pp. 123–131.
RUMPF, M. and STRZODKA, R. Graphics processor units: New prospects for parallel computing. In Are Magnus Bruaset and Aslak Tveito, editors, Numerical Solution of Partial Differential Equations on Parallel Computers, volume 51 of Lecture Notes in Computational Science and Engineering, pages 89–134. Springer, 2005
BOHN, C.-A. KOHONEN Feature Mapping Through Graphics Hardware. In Proceedings of 3rd Int. Conference on Computational Intelligence and Neurosciences 1998.